The CanDo (.cndo) file format, which was designed to describe DNA nanostructures, contains sufficient information to generate the all-atom models of these DNA nanostructures. This atomic model generator stores four standard reference atomic structures of basepairs A-T, T-A, G-C, and C-G. Each standard reference atomic structure consists of two phosphates, two deoxyriboses, and two paired bases. An all-atom model is created by placing the standard reference atomic structures, using rigid-body translation and rotation, to the positions and orientations of the basepairs given by the .cndo file format. We have developed a software that generates atomic models from .cndo files. The script is available here.
Users of this tool are kindly requested to cite the following reference:
- K Pan, DN Kim, F Zhang, MR Adendorff, H Yan, M Bathe. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nature Communications, 5: 5578 (2014). [ Pubmed Article ]
Notes.
- This atomic model generator requires MATLAB and UCSF Chimera.
- This atomic model generator does not consider conformational change of basepairs, sugars, and phosphates. Users may run the energy minimization script, molecular dynamics simulations, or other computations to simulate atomic-level details of DNA nanostructures.
Step 1. Add the two directories in the downloaded ZIP package to the MATLAB paths.
>> addpath cndo2atomic_v3.4
>> addpath topology2pdb_ver3_bulge
Step 2. Assign colors for the DNA strands. In this example, if a strand contains less than 100 nucleotides, then a color with the RGB value (190, 190, 190) is assigned to this strand. Otherwise a color with the RGB value (204, 121, 167) is assigned.
>> param.StrandColor = [190 190 190; 86 180, 233];
>> param.L_thres = 100;
Step 3. Assign the rendering resolution (typically between 3 and 6) and figure size (typically 640-by-480).
>> param.molmapResolution = 3;
>> param.WindowSize = [640 480];
Step 4. Assign the path to UCSF Chimera.
>> param.chimeraEXE = ‘”C:\Program Files\Chimera 1.8\bin\chimera.exe”‘;
>> param.chimeraOPTION = ‘––silent ––script’;
Step 5. Generate the atomic model
>> CAD_path = ‘example_4way_junction.cndo’;
>> work_DIR = ‘example_4way_junction’;
>> main_cndo2pdb(CAD_path, work_DIR, param);
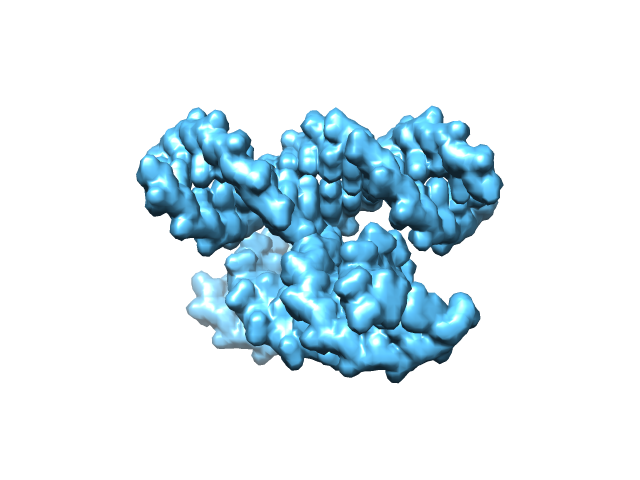
Finally, a directory ‘example_4way_junction’ is created in the current working directory. The files in the directory ‘example_4way_junction’ should be the same as those in example_4way_junction.zip. The generated atomic structure is shown below.
>> CAD_path = ‘tetrahedron.cndo’;
>> work_DIR = ‘tetrahedron’;
>> main_cndo2pdb(CAD_path, work_DIR, param);
A directory ‘tetrahedron’ is created in the current directory and is available here. The generated atomic structure is shown below.
>> CAD_path = ‘icosahedron.cndo’;
>> work_DIR = ‘icosahedron’;
>> main_cndo2pdb(CAD_path, work_DIR, param);
A directory ‘icosahedron’ is created in the current directory and is available here. The generated atomic structure is shown below.
>> CAD_path = ‘4_layer.cndo’;
>> work_DIR = ‘4_layer’;
>> main_cndo2pdb(CAD_path, work_DIR, param);
A directory ‘4_layer’ is created in the current directory and is available here. The generated atomic structure is shown below.
>> CAD_path = ‘hemisphere.cndo’;
>> work_DIR = ‘hemisphere’;
>> main_cndo2pdb(CAD_path, work_DIR, param);
A directory ‘hemisphere’ is created in the current directory and is available here. The generated atomic structure is shown below.