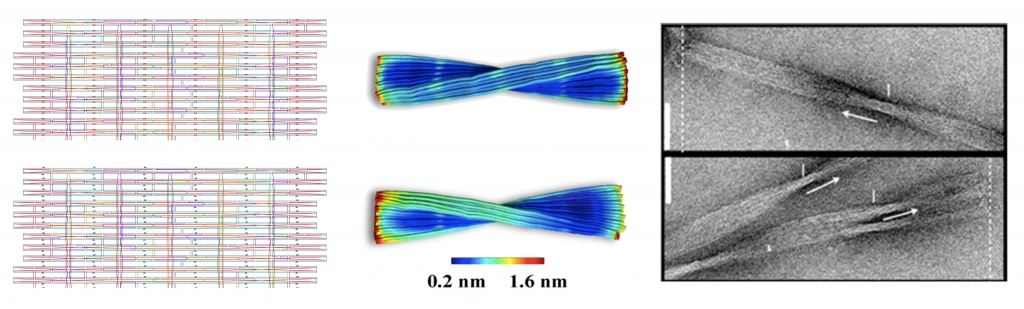
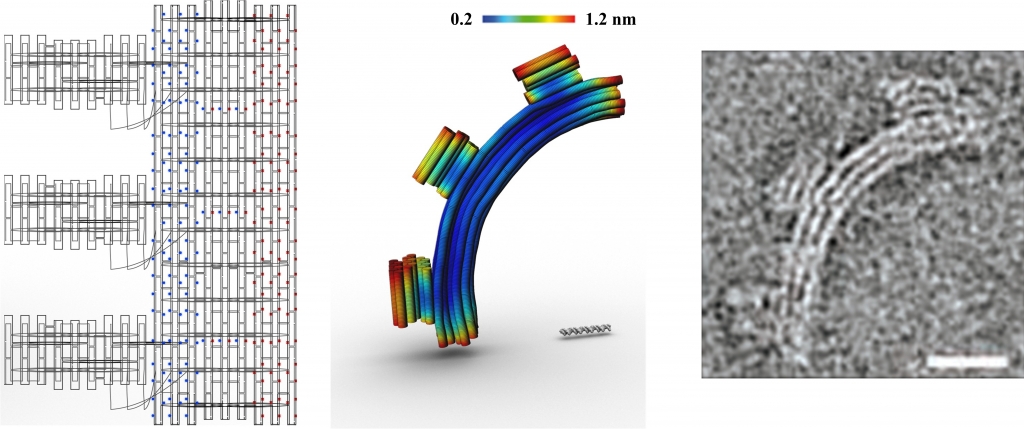
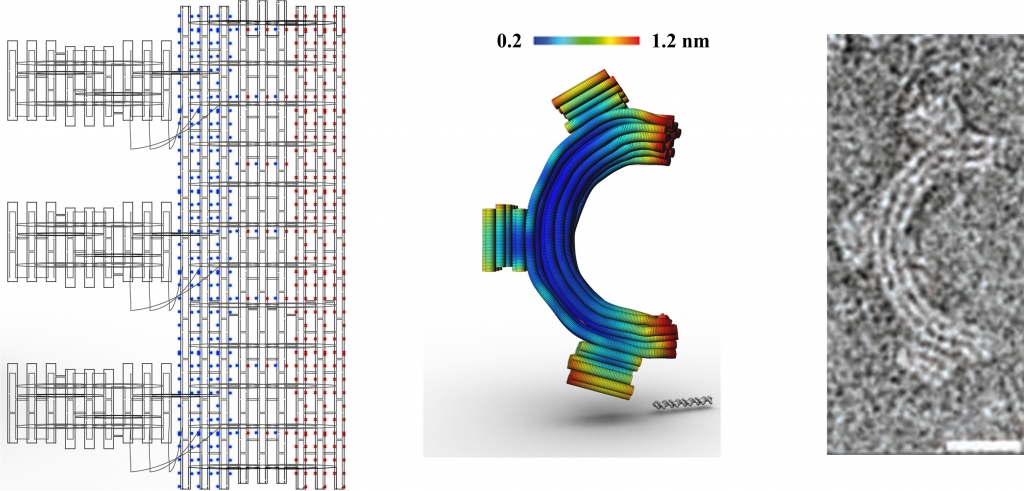
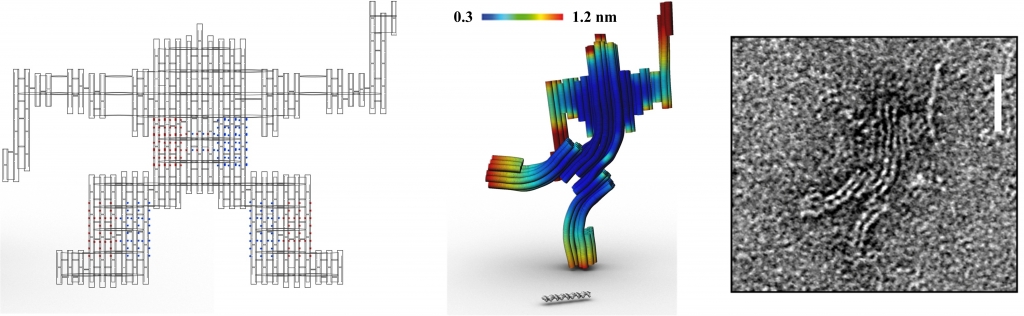
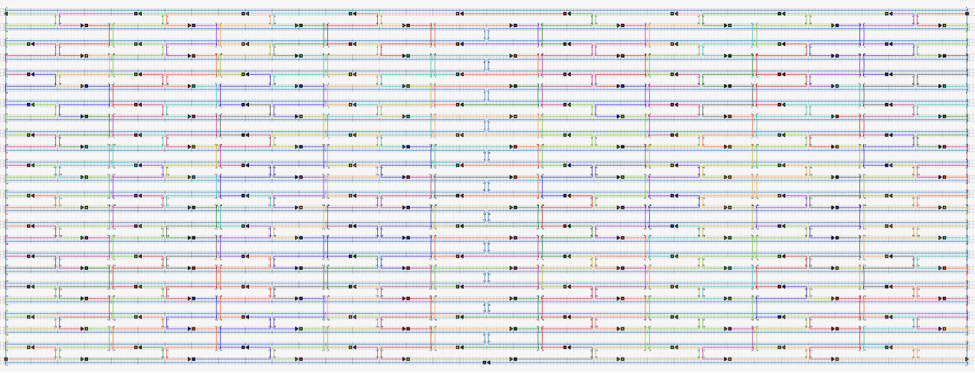
In Examples 1-4 below the planar caDNAno designs are shown at left, indicating the backbone scaffolds, crossovers, and insertions (blue dots) and deletions (red dots), followed by the 3D CanDo predicted shapes with flexibility indicated as heat-maps, where red and blue respectively indicates high and low flexibility, and finally the experimentally derived Transmission Electron Micrograph (TEM) images of the designed structures. Examples 5-6 provide .cndo design files that are fed into the atomic model generator to create the all-atom models. Examples 7-8 show Tiamat designs at left, followed by the all-atom models solved by CanDo, and finally the AFM (in example 7) and TEM (in example 8) images.
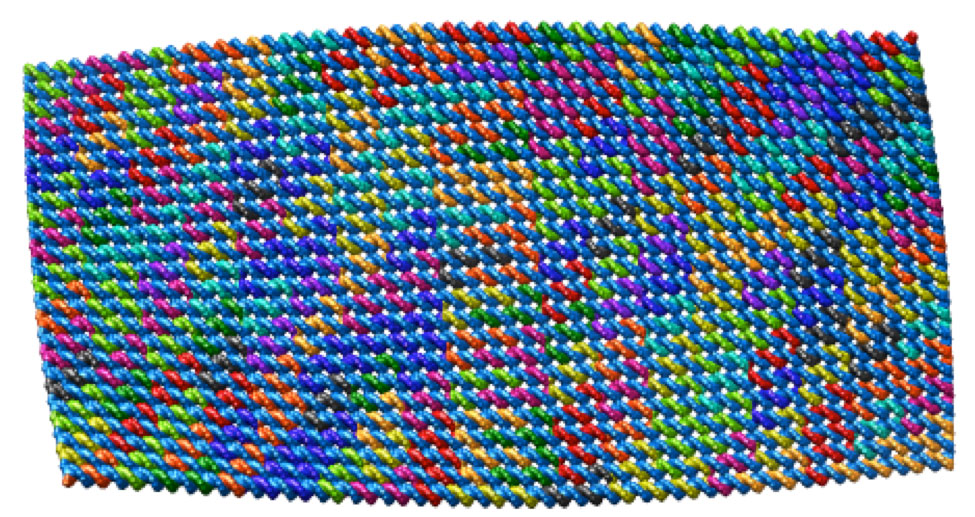
These twisted DNA-bundles [4] each consist of 10-by-6 DNA double helices constrained to a honeycomb lattice. The reference design, which is designed to remain straight, consists of 126 bp-long helices with 19 crossover planes evenly spaced every 7 bp along the helical axis, corresponding to 12 complete turns at 10.5 bp per turn. Two twisted versions of the reference design are realized by deleting and adding a single base-pair from every third cell array separating crossover planes, resulting in an overall left-handed (120 bp-long with 10 bp per turn) and right-handed (132 bp-long with 11 bp per turn) twist, respectively. The corresponding .json files for these designs can be downloaded here. Scale bars in the TEM images indicate 50 nm.
These DNA gears [4] are honeycomb-pleated 3-by-6 helix bundles that are designed to bend into either a quarter circle or a semi-circle with radii of 50 and 25 nm, respectively. Balanced gradients of insertions and deletions are used along the radial direction to produce global bend with no global twist. Neither insertions nor deletions are present in the tooth regions. The corresponding .json files for these designs can be downloaded here. Scale bars in the middle, FE model images are 50 base pairs. Scale bars in the TEM images indicate 20 nm.
This DNA sculpture of a robot is a 75 nanometer tall multi-layer DNA origami structure consisting of 74 double helices in a honeycomb lattice [7]. The DNA robot can be divided into three parts: body, arms, and legs, where balanced gradients of insertions and deletions are used in the body (waist) and two legs to induce curvature. The corresponding .json files for these designs can be downloaded here. Scale bar in the middle, FE model image is 50 base-pairs. Scale bar in the TEM image indicates 20 nm.
This DNA rectangle is a 50-by-100 nanometer single-layer DNA origami structure consisting of 24 double helices in a square lattice [2]. An atomic model of this DNA rectangle facilitates accurate design of megadalton-size nanodevices [14]. The corresponding .json file and .csv sequence file required by CanDo to construct the atomic model can be downloaded here.
This DNA tetrahedron is a wireframe DNA structure with the edge length of 18 nm. Each edge consists of two DNA duplexes. Due to the geometric shape, CanDo (.cndo) is a suitable file format for designing this DNA assembly. The CanDo design file can be downloaded here.

This DNA icosahedron is a wireframe DNA structure with the edge length of 18 nm. Each edge consists of two DNA duplexes. Due to the geometric shape, CanDo (.cndo) is a suitable file format for designing this DNA assembly. The CanDo design file can be downloaded here.

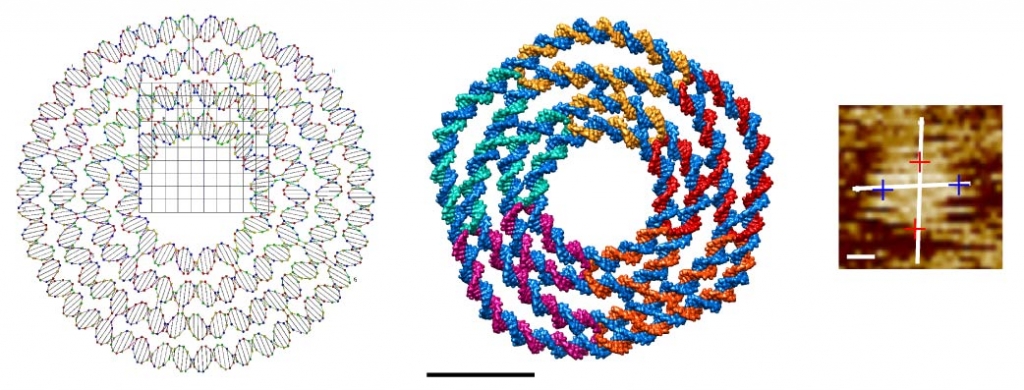
This DNA four-layer ring is a single-layer DNA structure with the inner diameter of 11 nm and the outer diameter of 27 nm. Curvature is introduced into each layer by controlling the number of base-pairs between neighboring four-way junctions. Due to the geometric shape, Tiamat is a suitable tool for designing this DNA assembly. The four layers consist of 100, 150, 200, and 250 base-pairs, respectively. The Tiamat (.dna) design file shown in the left panel can be downloaded here. Also available is the .cndo file corresponding to the atomic model shown in the middle panel. Scale bars in the atomic model and the AFM image are both 10 nm.
This DNA hemisphere is a single-layer DNA structure with the diameter of 42 nm. The design consists of 12 layers. Curvature is introduced into each layer by controlling the number of base-pairs between neighboring four-way junctions. Due to the geometric shape, Tiamat is a suitable tool for designing this DNA assembly. The Tiamat (.dna) design file shown in the left panel can be downloaded here. Also available is the .cndo file corresponding to the atomic model shown in the middle panel. Scale bars in the atomic model and the TEM images are both 10 nm.