User information provided will be used only for internal evaluation of the resource by counting the number of unique users and the number of unique affiliations. No other information will be stored on the server. Provided E-mail address will be used to send CanDo notifications when the analysis is queued, started, and completed. If not provided, no notification will be sent.
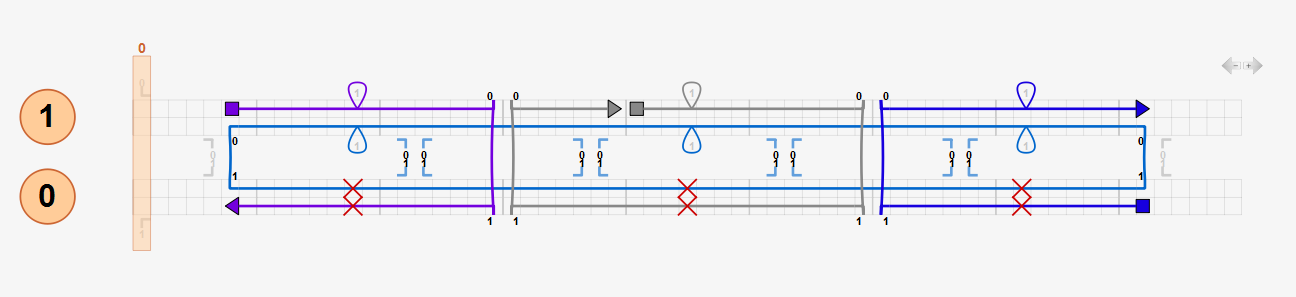
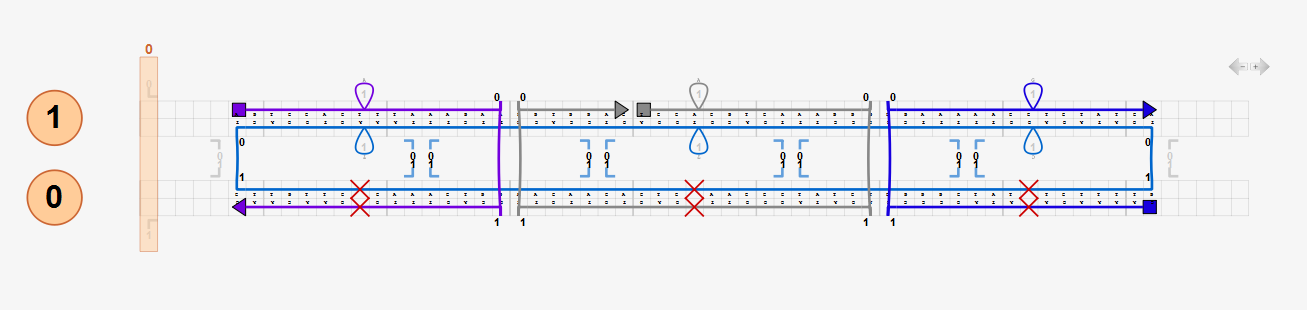
Default values for average B-form DNA mechanical properties are pre-entered. Users may enter their own values. Nicks are modeled by reducing backbone bending and torsional stiffness by a factor of 100 by default (corresponding to the default nick stiffness factor, 0.01) whereas stretching stiffness is retained at double-helix values. It is not recommended to use a nick stiffness factor less than the default value as it may result in much slower or no convergence of the analysis.
CanDo analysis is performed at the coarse model resolution by default. Users have an option to use the fine model resolution that computes the shape and flexibility at a single basepair resolution. However, the use of the coarse model is appropriate to obtain quick feedback for initial designs as it significantly reduces the computation time. Here we choose the fine resolution for purposes of this tutorial.
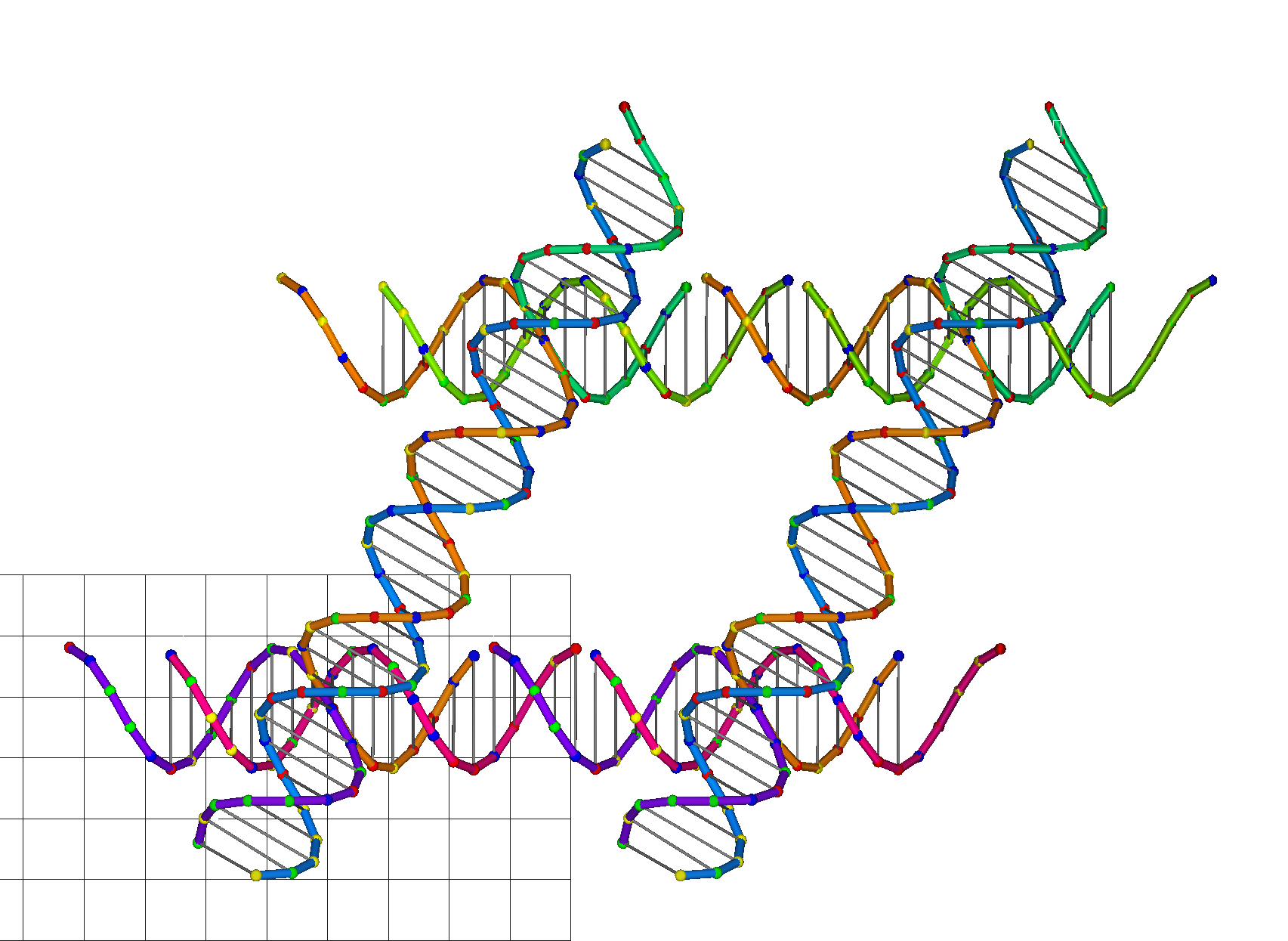
CanDo offers a new option for users to generate an atomic model of the solution shape of DNA nanostructure. Users can choose to generate an atomic model or compute the thermal fluctuations as well. CanDo analysis must be performed at the fine model resolution to generate the atomic model. If the atomic model is to be included in the results, make sure that the fine resolution is selected in step e. If thermal fluctuations of the atomic model are to be included, users must choose “Yes” in item h.
User information provided will be used only for internal evaluation of the resource by counting the number of unique users and the number of unique affiliations. No other information will be stored on the server. Provided E-mail address will be used to send CanDo notifications when the analysis is queued, started, and completed. If not provided, no notification will be sent.
Default values for average B-form DNA mechanical properties are pre-entered. Users may enter their own values. Nicks are modeled by reducing backbone bending and torsional stiffness by a factor of 100 by default (corresponding to the default nick stiffness factor, 0.01) whereas stretching stiffness is retained at double-helix values. It is not recommended to use a nick stiffness factor less than the default value as it may result in much slower or no convergence of the analysis.
CanDo offers a new option for users to generate an atomic model of the solution shape of DNA nanostructure. Users can choose to generate an atomic model or compute the thermal fluctuations as well. CanDo analysis must be performed at the fine model resolution to generate the atomic model. If thermal fluctuations of the atomic model are to be included, users must choose “Yes” in item f.